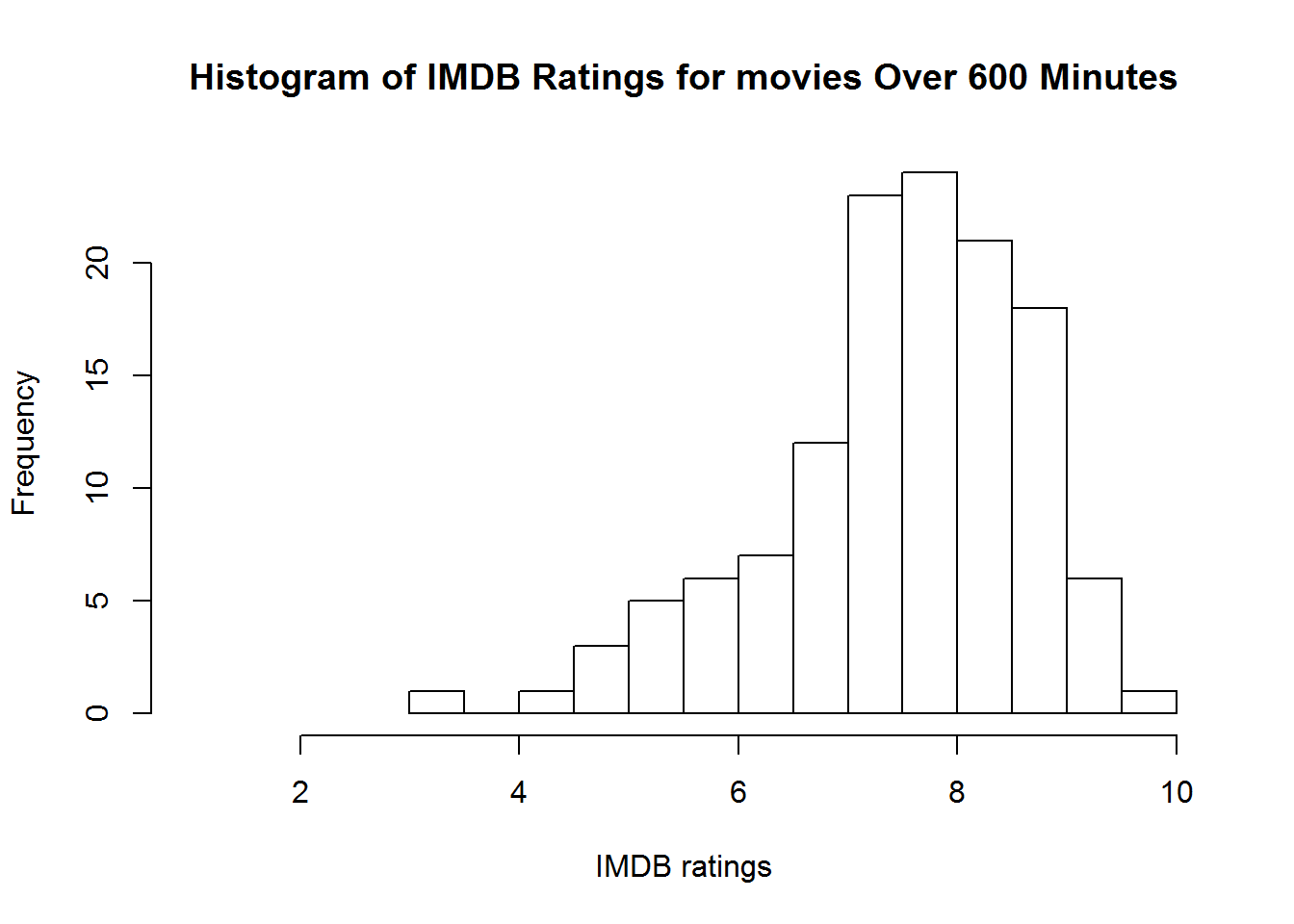Lesson 3: Intermediate Data Management
Scott Withrow
October, 07, 2015
Most of the time someone spends working on a data analysis problem will be getting the data into the program. This starts with reading in a file and turns into transforming variables, correcting mistakes, and sometimes redesigning the entire dataset (e.g., long to tall changes). In this module we will begin to talk about how we can manipulate data in R to be the form we want for the analyses we want to conduct.
We are going to begin working with an actual dataset that will hopefully be a little entertaining as well as informative. First, we set a working directory to save files, graphs, etc. into. We can also have R create directories for us Show warnings = FALSE ignores warnings telling us the directory already exists (if it does already exist)
dir.create("E:/Rcourse/L3", showWarnings = FALSE) Then set as working directory
setwd("E:/Rcourse/L3")The dataset we will be using is a from IMDB and Rotten Tomatoes. You can find and create your own dataset with these variables or use the one linked here. We can start off by saving the file to the directory we set in the setwd step.
download.file(“http://degovx.eurybia.feralhosting.com/movies.RData”, “orig_movies.RData”)
Note: Making directories close to a root or in a home directory (documents in Windows, home in Linux, and whatever the equivalent is on an Apple machine) will make things a little less messy. I tend to partition a drive for doing analyses that automatically backs up every couple of days.
Once we have saved our file we can load it with a simple load statement (for RData file). We will look at a few different types of loading formats over the rest of the course including csv and fixed widths. See lesson 1 for some examples of loading and how to get data from common statistical programs.
load("E:/Rcourse/L3/orig_movies.RData")The first step is to see how the data looks and to see how R has imported the various variables. We can do that with head (first 6 rows), tail (last 6 rows), summary (variable names and quantiles, max, and NAs of numeric variables), and str (type of variables and examples of the first few rows from each variable)
head(movies)## Title Year Appropriate Runtime Genre
## 1 Carmencita 1894 NOT RATED 1 min Documentary, Short
## 2 Le clown et ses chiens 1892 <NA> <NA> Animation, Short
## 3 Pauvre Pierrot 1892 <NA> 4 min Animation, Comedy, Short
## 4 Un bon bock 1892 <NA> <NA> Animation, Short
## 5 Blacksmith Scene 1893 UNRATED 1 min Short
## 6 Chinese Opium Den 1894 <NA> 1 min Short
## Released Director Writer Metacritic imdbRating
## 1 <NA> William K.L. Dickson <NA> NA 5.9
## 2 October 28, 1892 Émile Reynaud <NA> NA 6.5
## 3 October 28, 1892 Émile Reynaud <NA> NA 6.7
## 4 October 28, 1892 Émile Reynaud <NA> NA 6.6
## 5 May 09, 1893 William K.L. Dickson <NA> NA 6.3
## 6 October 17, 1894 William K.L. Dickson <NA> NA 5.9
## imdbVotes Language Country Awards rtomRating rtomMeter rtomVotes Fresh
## 1 982 <NA> USA <NA> NA NA NA NA
## 2 118 <NA> France <NA> NA NA NA NA
## 3 523 <NA> France <NA> NA NA NA NA
## 4 79 <NA> France <NA> NA NA NA NA
## 5 1134 <NA> USA 1 win. NA NA NA NA
## 6 53 English USA <NA> NA NA NA NA
## Rotten rtomUserMeter rtomUserRating rtomUserVotes BoxOffice
## 1 NA NA NA NA <NA>
## 2 NA NA NA NA <NA>
## 3 NA NA NA NA <NA>
## 4 NA 100 2.8 216 <NA>
## 5 NA 32 3.0 184 <NA>
## 6 NA NA NA NA <NA>summary(movies)## Title Year Appropriate
## Length:548010 Length:548010 Length:548010
## Class :character Class :character Class :character
## Mode :character Mode :character Mode :character
##
##
##
##
## Runtime Genre Released
## Length:548010 Length:548010 Length:548010
## Class :character Class :character Class :character
## Mode :character Mode :character Mode :character
##
##
##
##
## Director Writer Metacritic imdbRating
## Length:548010 Length:548010 Min. : 1.0 Min. : 1.00
## Class :character Class :character 1st Qu.: 45.0 1st Qu.: 5.60
## Mode :character Mode :character Median : 58.0 Median : 6.60
## Mean : 56.5 Mean : 6.38
## 3rd Qu.: 69.0 3rd Qu.: 7.30
## Max. :100.0 Max. :10.00
## NA's :537880 NA's :197639
## imdbVotes Language Country
## Min. : 5 Length:548010 Length:548010
## 1st Qu.: 11 Class :character Class :character
## Median : 29 Mode :character Mode :character
## Mean : 1614
## 3rd Qu.: 127
## Max. :1465210
## NA's :197640
## Awards rtomRating rtomMeter rtomVotes
## Length:548010 Min. : 0.0 Min. : 0.0 Min. : 1.0
## Class :character 1st Qu.: 5.0 1st Qu.: 40.0 1st Qu.: 6.0
## Mode :character Median : 6.2 Median : 67.0 Median : 14.0
## Mean : 6.0 Mean : 61.6 Mean : 34.8
## 3rd Qu.: 7.1 3rd Qu.: 86.0 3rd Qu.: 39.0
## Max. :10.0 Max. :100.0 Max. :328.0
## NA's :529206 NA's :529017 NA's :526140
## Fresh Rotten rtomUserMeter rtomUserRating
## Min. : 0.0 Min. : 0.0 Min. : 0.0 Min. :0.0
## 1st Qu.: 3.0 1st Qu.: 1.0 1st Qu.: 34.0 1st Qu.:2.2
## Median : 8.0 Median : 4.0 Median : 58.0 Median :3.1
## Mean : 21.6 Mean : 13.2 Mean : 55.8 Mean :2.6
## 3rd Qu.: 24.0 3rd Qu.: 12.0 3rd Qu.: 79.0 3rd Qu.:3.6
## Max. :311.0 Max. :195.0 Max. :100.0 Max. :5.0
## NA's :526140 NA's :526140 NA's :485854 NA's :455274
## rtomUserVotes BoxOffice
## Min. : 0 Length:548010
## 1st Qu.: 6 Class :character
## Median : 74 Mode :character
## Mean : 25271
## 3rd Qu.: 512
## Max. :35791395
## NA's :455131str(movies)## Classes 'tbl_df', 'tbl' and 'data.frame': 548010 obs. of 23 variables:
## $ Title : chr "Carmencita" "Le clown et ses chiens" "Pauvre Pierrot" "Un bon bock" ...
## $ Year : chr "1894" "1892" "1892" "1892" ...
## $ Appropriate : chr "NOT RATED" NA NA NA ...
## $ Runtime : chr "1 min" NA "4 min" NA ...
## $ Genre : chr "Documentary, Short" "Animation, Short" "Animation, Comedy, Short" "Animation, Short" ...
## $ Released : chr NA "October 28, 1892" "October 28, 1892" "October 28, 1892" ...
## $ Director : chr "William K.L. Dickson" "Émile Reynaud" "Émile Reynaud" "Émile Reynaud" ...
## $ Writer : chr NA NA NA NA ...
## $ Metacritic : int NA NA NA NA NA NA NA NA NA NA ...
## $ imdbRating : num 5.9 6.5 6.7 6.6 6.3 5.9 5.5 5.9 4.9 6.9 ...
## $ imdbVotes : int 982 118 523 79 1134 53 365 967 60 3376 ...
## $ Language : chr NA NA NA NA ...
## $ Country : chr "USA" "France" "France" "France" ...
## $ Awards : chr NA NA NA NA ...
## $ rtomRating : num NA NA NA NA NA NA NA NA NA NA ...
## $ rtomMeter : int NA NA NA NA NA NA NA NA NA NA ...
## $ rtomVotes : int NA NA NA NA NA NA NA NA NA NA ...
## $ Fresh : int NA NA NA NA NA NA NA NA NA NA ...
## $ Rotten : int NA NA NA NA NA NA NA NA NA NA ...
## $ rtomUserMeter : int NA NA NA 100 32 NA NA NA NA NA ...
## $ rtomUserRating: num NA NA NA 2.8 3 NA NA NA NA 3.8 ...
## $ rtomUserVotes : int NA NA NA 216 184 NA NA NA NA 124 ...
## $ BoxOffice : chr NA NA NA NA ...If you would like to interactively look at your dataset you can use view view(movies)
The first thing you should ask yourself is “How does the data look?” Then you should ask yourself “Are there any obvious issues?” Finally, you will probably have to ask yourself. How do I solve these problems?
For this dataset I see quite a few issues. Runtime, Released, Year, BoxOffice, Awards, and Appropriate are character vectors. Year and Released should be date variables. Genre, and Language have multiple entries in some rows. Genre, Language, Appropriate, Awards, and Country should all be nominal factors. Like most data analysis sessions getting the data into a format that is easy to use will be the first step. Followed by creating any variables that you may need. Using this data we will cover the basic tools of restructuring a dataset. ##Recoding Variables
Whether you are looking for a median split (shudder), you want to reverse code a scale, or your advisor has insisted you catagorize a continuous variable (because you would never do that yourself right?) you can do that relatively easily with base-R functionality. Recoding into a new variable is also great practice for the referencing we learned in previous lessons.
Let’s start out by breaking IMDB ratings down into bad (1-4), average (5-7), and good (8-10).
We can do this moderately quickly but cumbersomely with base-R First we duplicate the data from imdbRating into a new variable.
movies$imdbrate_cat <- movies$imdbRatingThen we take that new variable and we use the referencing to subset the variable into a smaller piece. Basically, we are saying from the movies dataset take the variable ($) imdbrate_cat. Then, within that variable find all instances where that variable meets a certain criterion. In this case that criterion is a logical operator. There are a number of simple logical operators in r.
Some Logical Operators:
- < less than
- <= less than or equal to
- > greater than
- >= greater than or equal to
- == exactly equal to
- != not equal to
- !x Not x
- x | y x OR y
- x & y x AND y
- isTRUE(x) test if X is TRUE
Behind the scenes R makes a vector of TRUE and FALSE statements for every row, then selects all the rows which equal TRUE. With that subset in mind we tell r to assign either “Good”, “Average”, or “Bad
movies$imdbrate_cat[movies$imdbrate_cat >= 8] <- "Good"
movies$imdbrate_cat[movies$imdbrate_cat > 4 &
movies$imdbrate_cat < 8] <- "Average"
movies$imdbrate_cat[movies$imdbrate_cat <= 4] <- "Bad"We can take a look at our catagorization with the table() function. Table creates a contingency table of counts of combinations of factor levels. See help(table) for more information.
table(movies$imdbrate_cat)##
## Average Bad Good
## 289971 21960 38440We should also summarize our new variable to see how it was created.
summary(movies$imdbrate_cat)## Length Class Mode
## 548010 character characterIt looks like this variable is still a character variable and needs to be transformed into a factor. See Lesson 1 if you don’t remember how.
movies$imdbrate_cat <- factor(movies$imdbrate_cat, ordered=TRUE,
levels=c("Bad", "Average", "Good"))
summary(movies$imdbrate_cat)## Bad Average Good NA's
## 21960 289971 38440 197639For many simple subsets or recodes this functionality is good. However, when you want to recode a lot of catagories at once using a package can be helpful. The best package for recoding (among a lot of other useful features that we will be using in this class) is the Companion to Applied Regression package.
require("car") #If you don't have it already install.packages("car")## Loading required package: carWe will be using the recode() function. For more help type ?car or ?recode
movies$imdbrate_cat <- NANA is missing data. R treats both numeric and character missing the same. you can find missing with is.na() and exclude missing with na.rm = TRUE or use listwise deletion with na.omit = TRUE
recode(variable, “character string of changes”, options) Here we are telling cat to recode imdb rating. We are giving it a character string that has a couple of commands that recode recognizes (hi and lo). For more information on the commands available for recode try help(recode). We are also telling it that the outcome of the recode should be a factor and that factor is ordinal by passing the levels argument. Levels works exactly like in the factor command in base-R.
movies$imdbrate_cat <- recode(movies$imdbRating, "8:hi='Good';lo:4='Bad';4:8='Average'",
as.factor.result = TRUE, levels=c("Bad", "Average", "Good"))
summary(movies$imdbrate_cat)## Bad Average Good NA's
## 21960 289971 38440 197639Later when we learn the apply statement we will learn how to recode any number of variables quickly using recode or base-R.
For now, let’s unload the car package since we won’t be using it anymore in this lesson.
detach("package:car", unload = TRUE)Renaming Variables
You can open an interactive window with fix() to rename or modify variables like you would in SPSS or Excel. However, I find the interactive window to be rather slow and clumsy to use for more than 1 or 2 variables at a time. There are ways to rename variables through base-R functionality but I find packages simplify things immensly. For this we will be using the package dplyr. We will be using this package extensively in the next data management section as well as through the rest of the course.
require(dplyr) # install.packages("dplyr")## Loading required package: dplyr
##
## Attaching package: 'dplyr'
##
## The following objects are masked from 'package:stats':
##
## filter, lag
##
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, unionAs you can see a few objects are “masked” from “package:Stats”. This means that those commands (filer, lag, intersect, etc.) will now be interpreted by the dplyr package rather than by the base-R package “Stats.” Normally, this won’t be much of an issue for most users since a well built package won’t break any functionality. However, this may not always be the case. If you find something just stops working try unloading any packages that aren’t essential to what you are doing. It can be a good tactic to load a package, do what you need with it, and unload it. Particularly, when you see one package masks commands from another package you will usually find functionality of one or both of the packages will be compromised. Pay attention to this warnings and if you want to use one of the commands that is being masked look into how that new command works to make sure you aren’t accidently making errors that can hinder interpretation of your results later.
rename(dataset, new_variable_name = old_variable_name). Additional variables can be added seperated by a comma.
movies <- rename(movies, imdbRatingCatagory = imdbrate_cat)Date Variables
We have a couple of date variables in the dataset that will need to have some work done on them but the released variable needs the most so let’s start there. First, let’s take a look at released.
summary(movies$Released)## Length Class Mode
## 548010 character characterstr(movies$Released)## chr [1:548010] NA "October 28, 1892" "October 28, 1892" ...head(movies$Released)## [1] NA "October 28, 1892" "October 28, 1892"
## [4] "October 28, 1892" "May 09, 1893" "October 17, 1894"We have a character variable with the form MMMM dd, yyyy and some missing data. To see how R works with character data try visualing the variable with plot(movies$Released)
Looks like a character variable is unplottable. You could try a few other commands but you would find that character variables are pretty useless for analysis purposes. Later, we will find some uses for character variables, but most of the time you will want to get variables in the form of a number or factor.
R lets us tell it how variables should be classified with as. For example as.date(), as.numeric(), as.vector(), as.data.frame(), as.logical(). We can also test typing with is. for example, is.date() or is.numeric().
Before we convert this character string into a date we should make sure our localization is set correctly. In this case the data are English United States format. If your computer isn’t set to that localization you will have a bad time dealing with these dates.
Luckily, we can set localizations eazily.
Sys.setlocale("LC_TIME", "English")## [1] "English_United States.1252"Let’s try setting this variable as a date
head(as.Date(movies$Released))
The R as.Date command is well scripted and can recognize most common date formats. Sometimes, like above, you might run into something a little strange or unusual and you might need to set the format yourself. You can do that pretty easily. R uses POSIX for conversion. To find the commands specific to your format try help(strptime).
Looking at the help file we can see that a full month written out is a %B while the day is %d and four digit year is %Y
Note: as.Date uses a standard representation of time which sets a specific second as reference and all dates and times are stored as the time since or before that point.
We can specify our format as well as overwriting the current variable by giving as.Date a character string with the exact format (including commas, slashes, and dashes). We can also specify the time zone if there are hours, minutes, seconds, etc. in the string.
movies$Released <- as.Date(movies$Released, "%B %d, %Y", tz = "UTC")
summary(movies$Released)## Min. 1st Qu. Median Mean 3rd Qu.
## "1888-10-14" "1982-12-03" "2004-09-23" "1993-04-16" "2010-06-13"
## Max. NA's
## "2022-01-01" "66665"str(movies$Released)## Date[1:548010], format: NA "1892-10-28" "1892-10-28" "1892-10-28" "1893-05-09" ...head(movies$Released)## [1] NA "1892-10-28" "1892-10-28" "1892-10-28" "1893-05-09"
## [6] "1894-10-17"Now our file has an appropriate date variable. We can see from summary that we are dealing with movies as early as 1888 and as late as 2022 (projected movies).
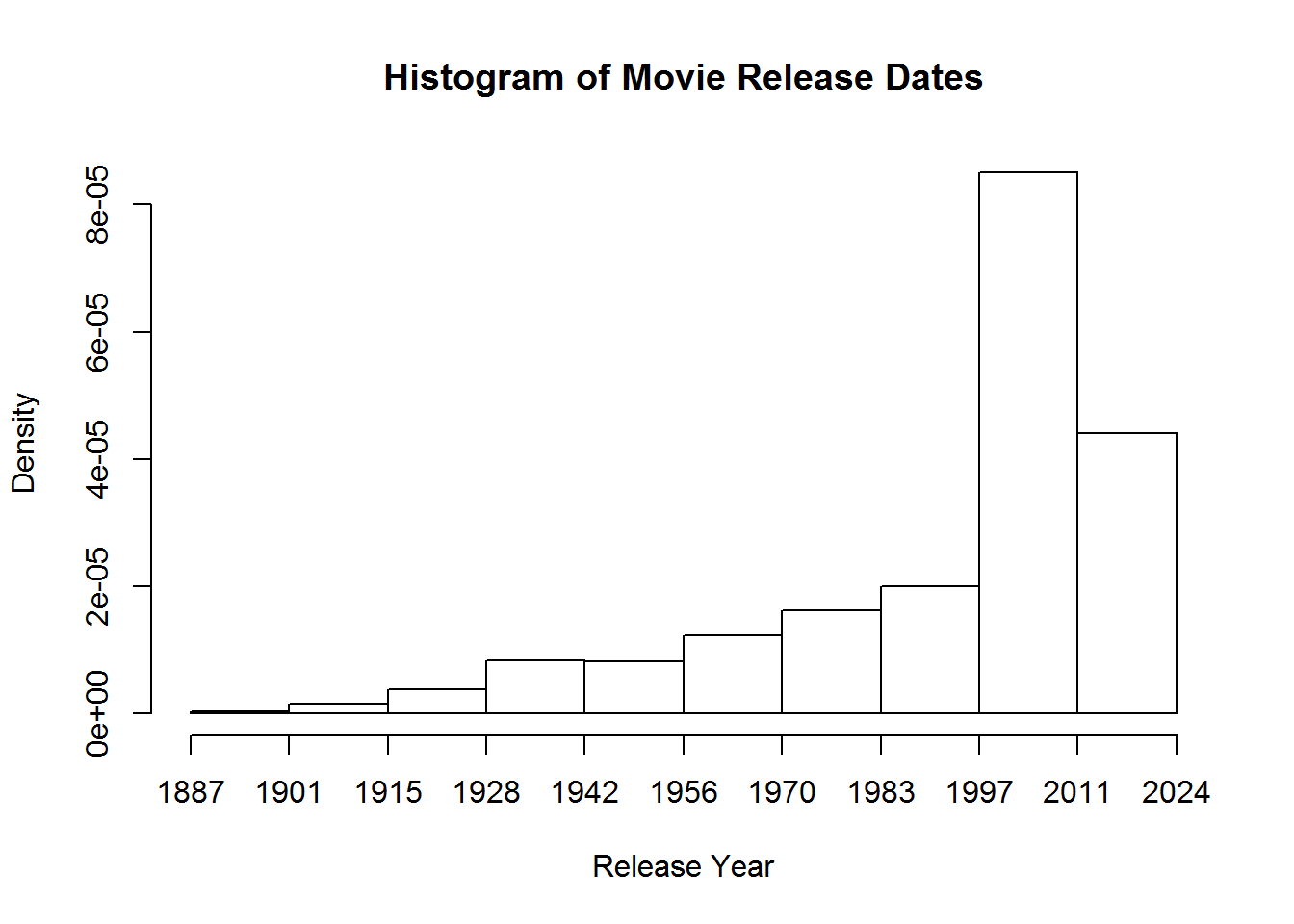
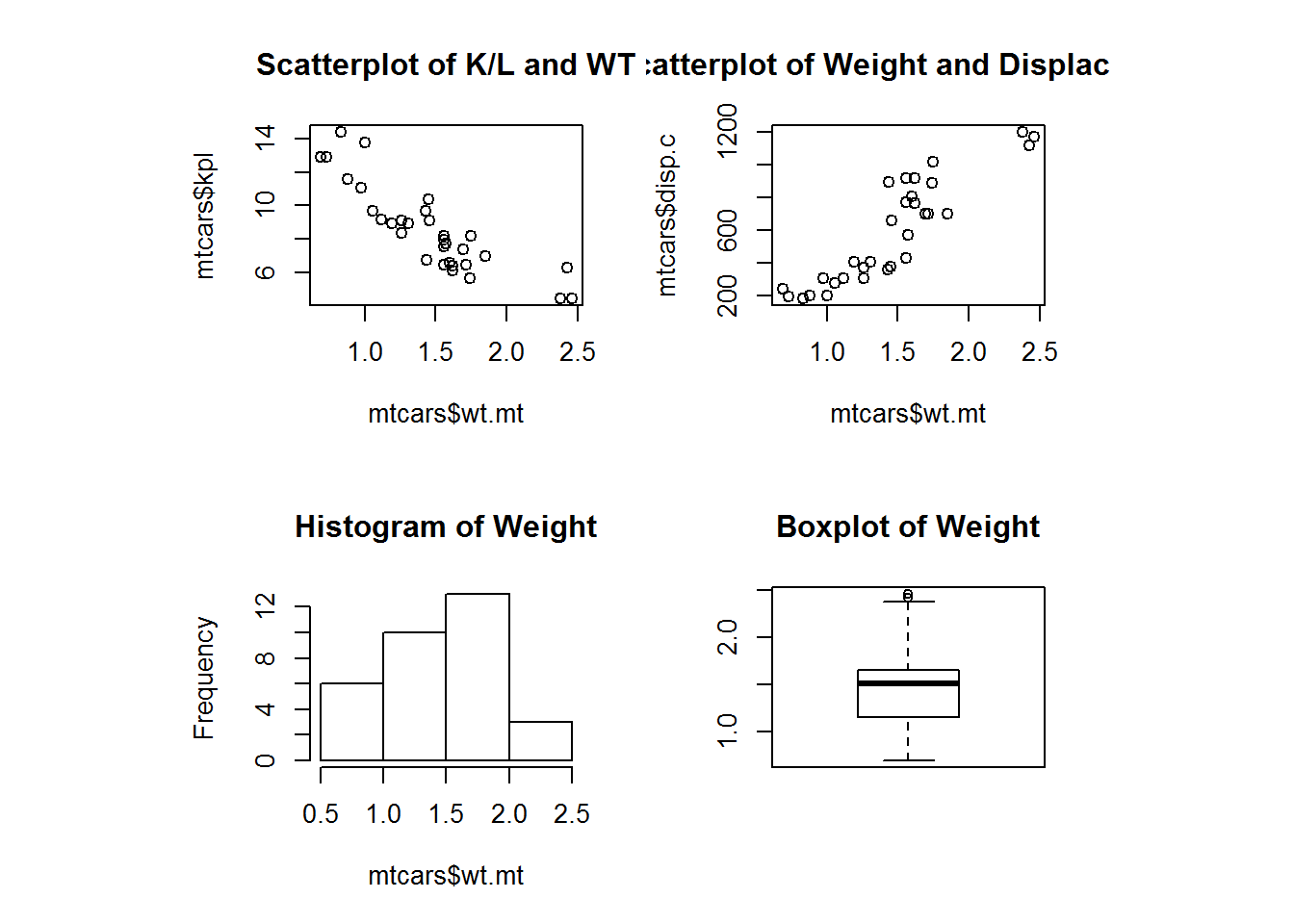
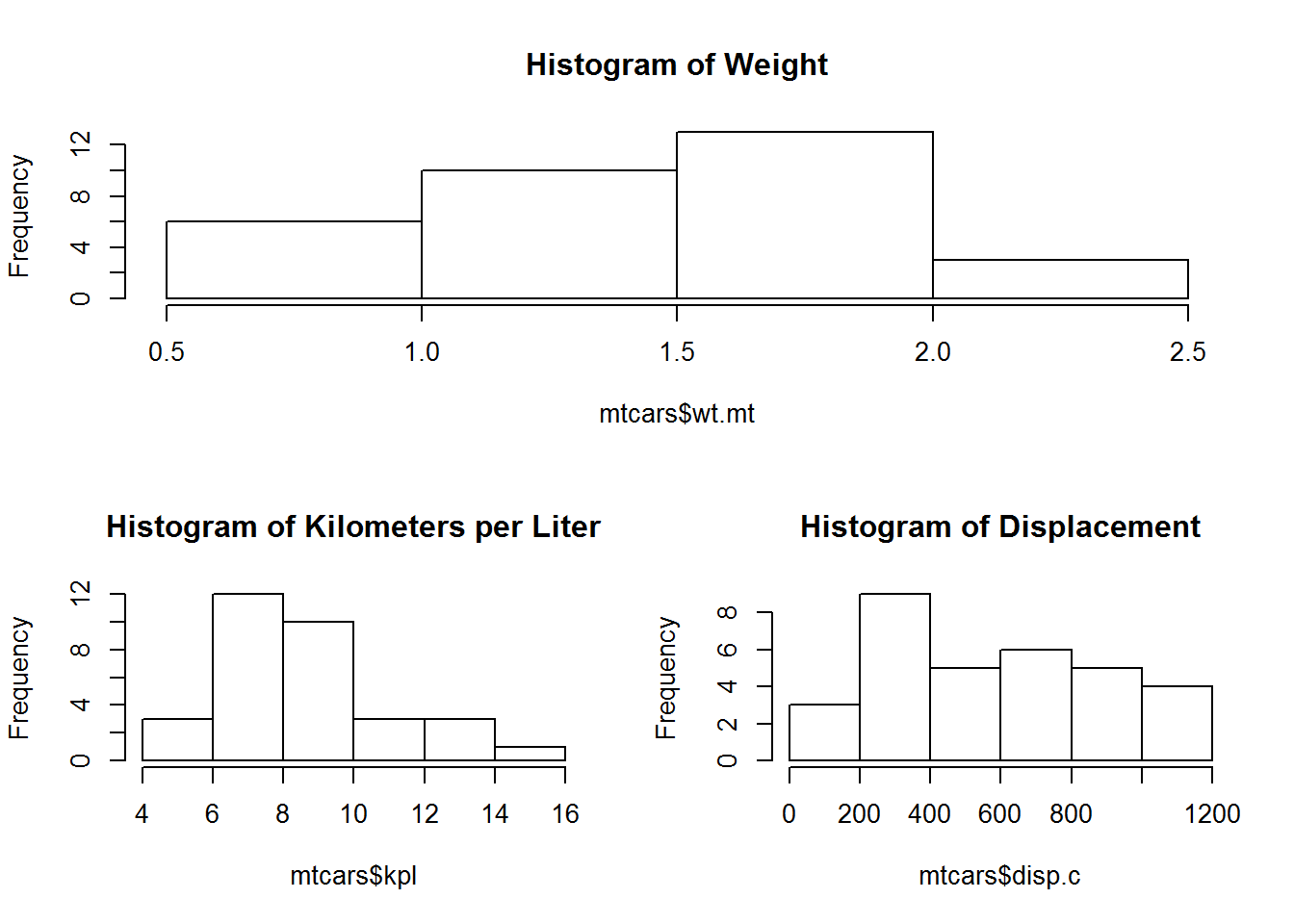
We can create a quick graph to look at number of movies released over time
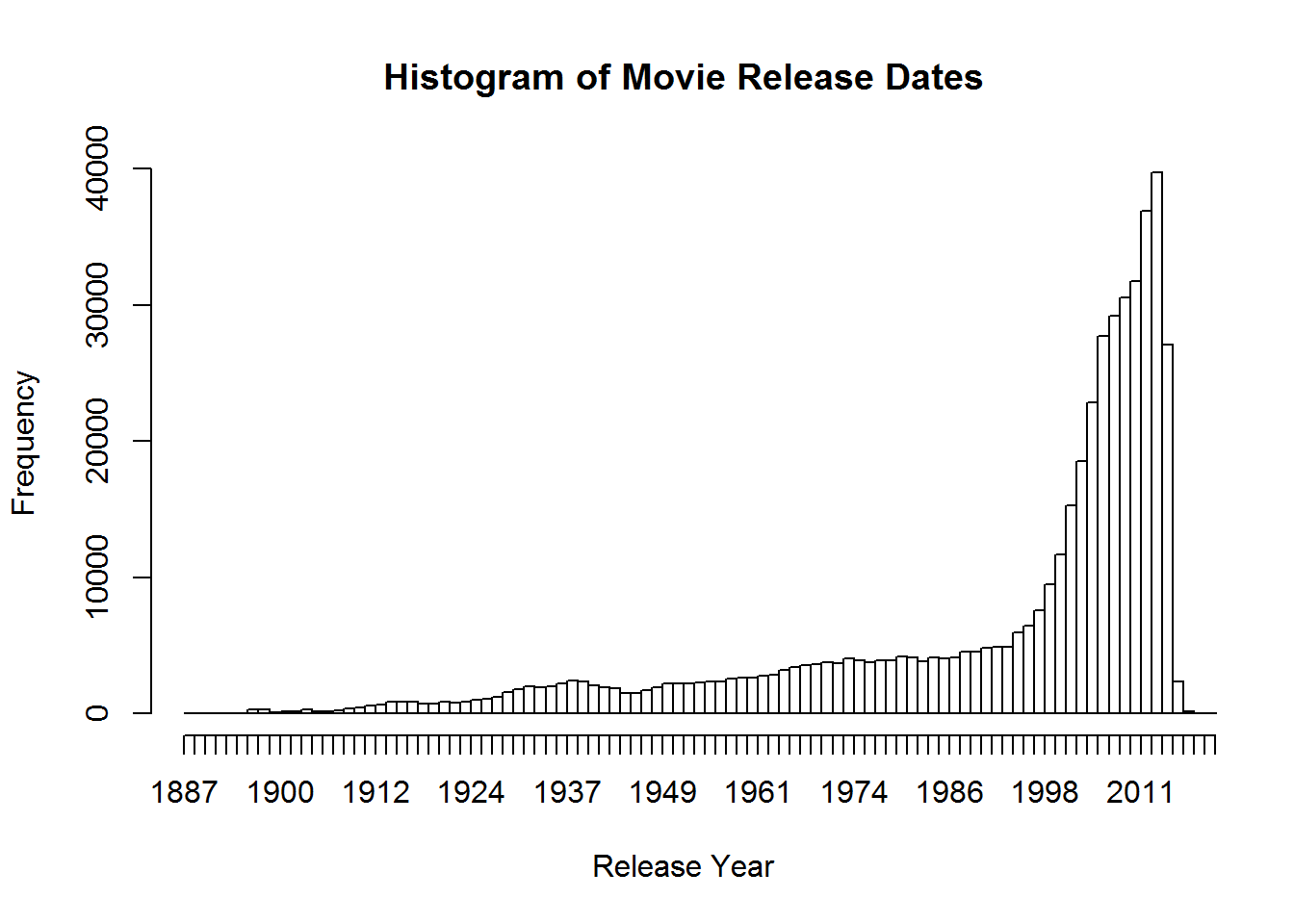
hist(movies$Released, breaks = 10, main="Histogram of Movie Release Dates", xlab="Release Year")Using some of the fancy things we learned from last lesson we can put together a better graph. Let’s break it up so this histogram has a break for every year, change from density to frequency, and cut off the projected movies from the graph. We will have to specify that we want the x axis to be in years by using the as.Date command and format. Format will let us specify how we want the date represented. In this case, we want just years.
hist(movies$Released, breaks = (2015 - 1888), freq = TRUE,
main = "Histogram of Movie Release Dates", xlab = "Release Year",
xlim = c(as.Date("1888", format="%Y"), as.Date("2015", format="%Y")))Regular Expressions in R
Now let’s convert Runtime to numeric. As we can see it’s a character vector still.
summary(movies$Runtime)## Length Class Mode
## 548010 character characterstr(movies$Runtime)## chr [1:548010] "1 min" NA "4 min" NA "1 min" "1 min" ...head(movies$Runtime)## [1] "1 min" NA "4 min" NA "1 min" "1 min"We can try using the as.numberic statement like we did with as.Date
head(as.numeric(movies$Runtime))## Warning in head(as.numeric(movies$Runtime)): NAs introduced by coercion## [1] NA NA NA NA NA NAUhoh looks like R can’t handle that min in the string.
We can get around that by using pattern matching on the data. In this case we can use gsub (grep substitution of all matches using regular expressions) Use ?gsub for more information. Regular expressions are very powerful and spending some time learning how to use them will be a great benefit not only to R programming but any use of computers. You can learn a lot about regex from Wikipedia (https://en.wikipedia.org/wiki/Regular_expression) or from http://www.regular-expressions.info/tutorial.html
In this case we need only the most basic and simple search to accomplish our goals.
movies$Runtime <- as.numeric(gsub(" min", "", movies$Runtime))## Warning: NAs introduced by coercionsummary(movies$Runtime)## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 0.15 19.00 60.00 61.10 91.00 964.00 76827str(movies$Runtime)## num [1:548010] 1 NA 4 NA 1 1 1 1 45 1 ...head(movies$Runtime)## [1] 1 NA 4 NA 1 1Alright, we now have a usable variable. We can see we have movies from 15 seconds long to a little over 16 hours long. “Wait, what?” you are saying. 16 hour long movies? We can learn a bit about that exceptionally long movie fairly easily. We can find the maximum number with max() and remove NA’s from the analysis with na.rm = TRUE
max(movies$Runtime, na.rm = TRUE)## [1] 964Then all we have to do is look up that value. The easiest way to do that is to use which(). Which takes a logical vector (a vector of true and false statements) and returns the row number(s) that match the logic. Which is a useful function since it automatically allows NAs and sets them to FALSE. This is important because it gives an accurate return index. It’s a useful shortcut to a long statement.
movies[which(movies$Runtime == 964), ]## Source: local data frame [1 x 24]
##
## Title Year Appropriate Runtime Genre Released
## (chr) (chr) (chr) (dbl) (chr) (date)
## 1 24 in Seven 2009 NA 964 Documentary, Adventure 2009-08-01
## Variables not shown: Director (chr), Writer (chr), Metacritic (int),
## imdbRating (dbl), imdbVotes (int), Language (chr), Country (chr), Awards
## (chr), rtomRating (dbl), rtomMeter (int), rtomVotes (int), Fresh (int),
## Rotten (int), rtomUserMeter (int), rtomUserRating (dbl), rtomUserVotes
## (int), BoxOffice (chr), imdbRatingCatagory (fctr)“24 in Seven” is the long movie that doesn’t have any ratings. We can use which to find other things too. Like movies that are longer than 10 hours. Here we can use the index to select only titles and ratings.
movies[which(movies$Runtime > 600), c("Title", "imdbRating")]## Source: local data frame [167 x 2]
##
## Title imdbRating
## (chr) (dbl)
## 1 Semnadtsat mgnoveniy vesny 9.1
## 2 War & Peace 8.2
## 3 Chlopi 7.2
## 4 Noce i dnie 7.4
## 5 I, Claudius 9.1
## 6 How Yukong Moved the Mountains 7.2
## 7 My Uncle Napoleon 8.6
## 8 Washington: Behind Closed Doors 8.0
## 9 Die Buddenbrooks 8.1
## 10 Flambards 8.3
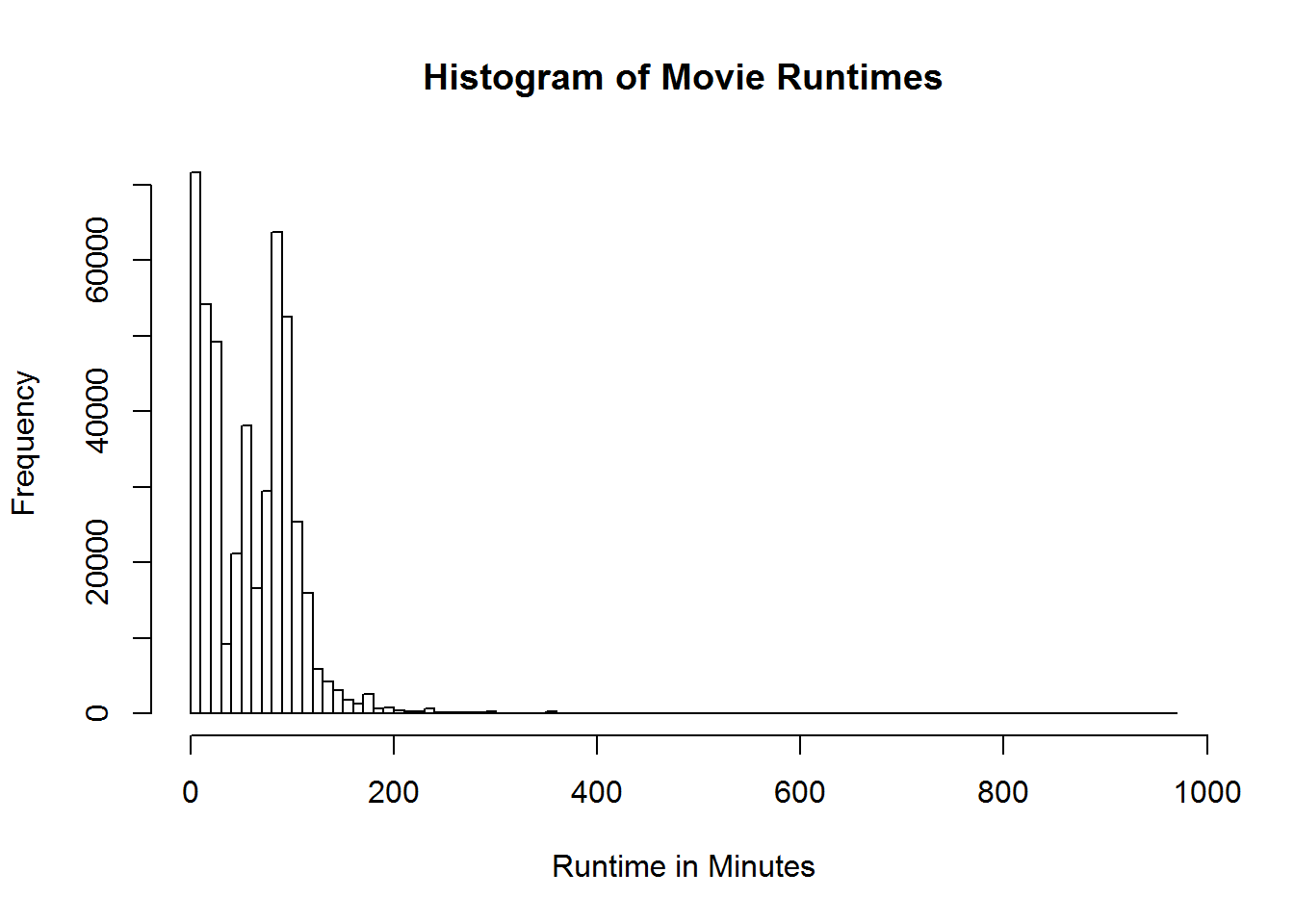
## .. ... ...Let’s use our graphing skills again
hist(movies$Runtime, breaks = 100, main = "Histogram of Movie Runtimes",
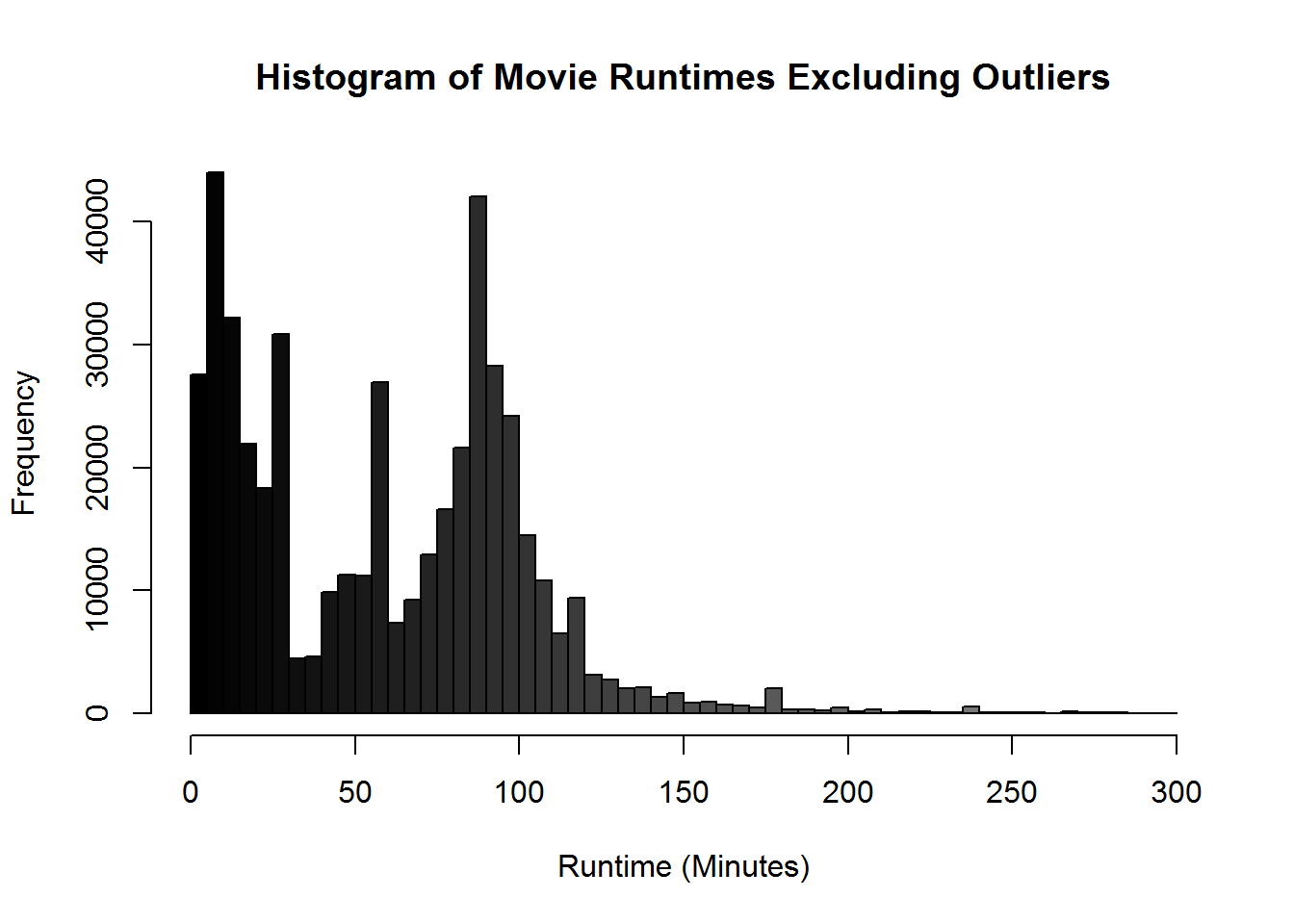
xlab = "Runtime in Minutes")Those pesky outliers are making our graph pretty hard to read. Let’s drop down the graph to be movies that are just 5 hours or less. Before we just modified the graph to exclude those values. Let’s use the subsetting we learned to reduce the actual data being processed by the graph. This will generally be the prefered way since it speeds up the rest of the graphing process and takes less computational cycles.
hist(movies$Runtime[movies$Runtime < 300], breaks = 100,
main = "Histogram of Movie Runtimes Excluding Outliers",
col = gray(0:100/100), xlab = "Runtime (Minutes)")View the data again now that we have transformed it.
summary(movies)## Title Year Appropriate Runtime
## Length:548010 Length:548010 Length:548010 Min. : 0.15
## Class :character Class :character Class :character 1st Qu.: 19.00
## Mode :character Mode :character Mode :character Median : 60.00
## Mean : 61.10
## 3rd Qu.: 91.00
## Max. :964.00
## NA's :76827
## Genre Released Director
## Length:548010 Min. :1888-10-14 Length:548010
## Class :character 1st Qu.:1982-12-03 Class :character
## Mode :character Median :2004-09-23 Mode :character
## Mean :1993-04-16
## 3rd Qu.:2010-06-13
## Max. :2022-01-01
## NA's :66665
## Writer Metacritic imdbRating imdbVotes
## Length:548010 Min. : 1.0 Min. : 1.00 Min. : 5
## Class :character 1st Qu.: 45.0 1st Qu.: 5.60 1st Qu.: 11
## Mode :character Median : 58.0 Median : 6.60 Median : 29
## Mean : 56.5 Mean : 6.38 Mean : 1614
## 3rd Qu.: 69.0 3rd Qu.: 7.30 3rd Qu.: 127
## Max. :100.0 Max. :10.00 Max. :1465210
## NA's :537880 NA's :197639 NA's :197640
## Language Country Awards rtomRating
## Length:548010 Length:548010 Length:548010 Min. : 0.0
## Class :character Class :character Class :character 1st Qu.: 5.0
## Mode :character Mode :character Mode :character Median : 6.2
## Mean : 6.0
## 3rd Qu.: 7.1
## Max. :10.0
## NA's :529206
## rtomMeter rtomVotes Fresh Rotten
## Min. : 0.0 Min. : 1.0 Min. : 0.0 Min. : 0.0
## 1st Qu.: 40.0 1st Qu.: 6.0 1st Qu.: 3.0 1st Qu.: 1.0
## Median : 67.0 Median : 14.0 Median : 8.0 Median : 4.0
## Mean : 61.6 Mean : 34.8 Mean : 21.6 Mean : 13.2
## 3rd Qu.: 86.0 3rd Qu.: 39.0 3rd Qu.: 24.0 3rd Qu.: 12.0
## Max. :100.0 Max. :328.0 Max. :311.0 Max. :195.0
## NA's :529017 NA's :526140 NA's :526140 NA's :526140
## rtomUserMeter rtomUserRating rtomUserVotes BoxOffice
## Min. : 0.0 Min. :0.0 Min. : 0 Length:548010
## 1st Qu.: 34.0 1st Qu.:2.2 1st Qu.: 6 Class :character
## Median : 58.0 Median :3.1 Median : 74 Mode :character
## Mean : 55.8 Mean :2.6 Mean : 25271
## 3rd Qu.: 79.0 3rd Qu.:3.6 3rd Qu.: 512
## Max. :100.0 Max. :5.0 Max. :35791395
## NA's :485854 NA's :455274 NA's :455131
## imdbRatingCatagory
## Bad : 21960
## Average:289971
## Good : 38440
## NA's :197639
##
##
## It looks like we still need to work on the variables Year, Appropriate, Genre, Director, Writer, Language, Country, Awards, and BoxOffice.
We will want to use Genre so let’s start there. First, we can use table to see a better summary of the data.
head(table(movies$Genre))##
## Action Action, Adult Action, Adult, Adventure
## 4563 14 6
## Action, Adult, Animation Action, Adult, Comedy Action, Adult, Crime
## 1 8 5Here, with just head we can see that many catagories are present seperated by a comma. We could use regex to seperate these into new catagories but luckily for us there are already packages that can accomplish splitting, stacking, and shaping. Extra luckily for us these strings of combined information are constructed so the first entry is most important, second most, etc. That means we can take the first few catagories for each of these splits.
To accomplish the splitting we will be using tidyr. This is part of the family of packages like dplyr and plyr that we will be using in this course to manage data. Here we will be using a basic functionality to seperate a column of data into a number of new columns.
require(tidyr) # install.packages("tidyr")## Loading required package: tidyrthe command we are interested in is separate(). separate(data, col, into, sep = “[^[:alnum:]]+”, remove = TRUE, convert = FALSE, extra = “error”, …)
For our purposes we are going to use the data movies, the column Genre, we want to make three new columns Genre_1, Genre_2, Genre_3, keep the original column in the data, split based on a comma, and drop any extra columns of data after three.
movies_genresplit <- separate(movies, Genre, c("Genre_1", "Genre_2", "Genre_3"),
remove = FALSE, sep = ",", extra = "drop")tidyr is generally quick and easy but there are alternatives. A good alternative is the splitstackshape package which will automaticaly create a number of columns equal to the maximum number of splits. Splitstackshape uses the package data.table which we will not be covering in this course so once it has been used the resulting data needs to be converted back into a data frame before we can use it.
First, we will have to unload some packages since they conflict with splitstackshape
detach("package:dplyr", unload = TRUE)
detach("package:tidyr", unload = TRUE)splitstackshape example
require("splitstackshape")## Loading required package: splitstackshape
## Loading required package: data.tablemovies <- cSplit(as.data.frame(movies), "Genre", sep=",")
setDF(movies)
summary(movies)## Title Year Appropriate Runtime
## Length:548010 Length:548010 Length:548010 Min. : 0.15
## Class :character Class :character Class :character 1st Qu.: 19.00
## Mode :character Mode :character Mode :character Median : 60.00
## Mean : 61.10
## 3rd Qu.: 91.00
## Max. :964.00
## NA's :76827
## Released Director Writer
## Min. :1888-10-14 Length:548010 Length:548010
## 1st Qu.:1982-12-03 Class :character Class :character
## Median :2004-09-23 Mode :character Mode :character
## Mean :1993-04-16
## 3rd Qu.:2010-06-13
## Max. :2022-01-01
## NA's :66665
## Metacritic imdbRating imdbVotes Language
## Min. : 1.0 Min. : 1.00 Min. : 5 Length:548010
## 1st Qu.: 45.0 1st Qu.: 5.60 1st Qu.: 11 Class :character
## Median : 58.0 Median : 6.60 Median : 29 Mode :character
## Mean : 56.5 Mean : 6.38 Mean : 1614
## 3rd Qu.: 69.0 3rd Qu.: 7.30 3rd Qu.: 127
## Max. :100.0 Max. :10.00 Max. :1465210
## NA's :537880 NA's :197639 NA's :197640
## Country Awards rtomRating rtomMeter
## Length:548010 Length:548010 Min. : 0.0 Min. : 0.0
## Class :character Class :character 1st Qu.: 5.0 1st Qu.: 40.0
## Mode :character Mode :character Median : 6.2 Median : 67.0
## Mean : 6.0 Mean : 61.6
## 3rd Qu.: 7.1 3rd Qu.: 86.0
## Max. :10.0 Max. :100.0
## NA's :529206 NA's :529017
## rtomVotes Fresh Rotten rtomUserMeter
## Min. : 1.0 Min. : 0.0 Min. : 0.0 Min. : 0.0
## 1st Qu.: 6.0 1st Qu.: 3.0 1st Qu.: 1.0 1st Qu.: 34.0
## Median : 14.0 Median : 8.0 Median : 4.0 Median : 58.0
## Mean : 34.8 Mean : 21.6 Mean : 13.2 Mean : 55.8
## 3rd Qu.: 39.0 3rd Qu.: 24.0 3rd Qu.: 12.0 3rd Qu.: 79.0
## Max. :328.0 Max. :311.0 Max. :195.0 Max. :100.0
## NA's :526140 NA's :526140 NA's :526140 NA's :485854
## rtomUserRating rtomUserVotes BoxOffice imdbRatingCatagory
## Min. :0.0 Min. : 0 Length:548010 Bad : 21960
## 1st Qu.:2.2 1st Qu.: 6 Class :character Average:289971
## Median :3.1 Median : 74 Mode :character Good : 38440
## Mean :2.6 Mean : 25271 NA's :197639
## 3rd Qu.:3.6 3rd Qu.: 512
## Max. :5.0 Max. :35791395
## NA's :455274 NA's :455131
## Genre_1 Genre_2 Genre_3 Genre_4
## Short :110902 Drama : 62760 Drama : 15207 Thriller: 269
## Documentary: 86989 Short : 46557 Romance : 11662 Romance : 246
## Drama : 81578 Comedy : 36044 Thriller: 9668 Family : 200
## Comedy : 74449 Romance: 14718 Comedy : 7354 Drama : 165
## Action : 25638 Crime : 9770 Family : 6791 Fantasy : 151
## (Other) :131529 (Other):102559 (Other) : 50743 (Other) : 1045
## NA's : 36925 NA's :275602 NA's :446585 NA's :545934
## Genre_5 Genre_6 Genre_7 Genre_8
## Thriller: 80 Thriller: 16 Sci-Fi : 5 Music : 1
## Romance : 52 Romance : 14 Thriller: 4 Musical : 1
## Sci-Fi : 45 Sci-Fi : 6 Music : 2 Romance : 1
## War : 40 Sport : 6 War : 2 Sci-Fi : 1
## Fantasy : 33 Mystery : 5 Adult : 1 Thriller: 4
## (Other) : 210 (Other) : 25 (Other) : 4 War : 1
## NA's :547550 NA's :547938 NA's :547992 NA's :548001Using either method we have sucessfully broken the Genre variable into multiple groups. Depending on which method we use we will have to do some cleanup. Using tidyr we would need to set the variables as factors since they are still strings. Using splitstackshape we can delete the extra columns that aren’t providing much information. For now, let’s use the output from splitstackshape and cover how to delete columns in R.
Subsetting a Dataset
It looks like we got 8 genre categories although the 4th genre only contains a handful of entries and the 8th only has 9. Let’s go ahead and delete the Genre_4 through 8. We can keep or drop variables a number of different ways.
Like we did above in the recoding section we can use dataframe element selection.
Since the extra columns are at the end of the dataset we can keep the beginning.
head(movies[, 1:26]) #We could just set this as movies with <-## Title Year Appropriate Runtime Released
## 1 Carmencita 1894 NOT RATED 1 <NA>
## 2 Le clown et ses chiens 1892 <NA> NA 1892-10-28
## 3 Pauvre Pierrot 1892 <NA> 4 1892-10-28
## 4 Un bon bock 1892 <NA> NA 1892-10-28
## 5 Blacksmith Scene 1893 UNRATED 1 1893-05-09
## 6 Chinese Opium Den 1894 <NA> 1 1894-10-17
## Director Writer Metacritic imdbRating imdbVotes Language
## 1 William K.L. Dickson <NA> NA 5.9 982 <NA>
## 2 Émile Reynaud <NA> NA 6.5 118 <NA>
## 3 Émile Reynaud <NA> NA 6.7 523 <NA>
## 4 Émile Reynaud <NA> NA 6.6 79 <NA>
## 5 William K.L. Dickson <NA> NA 6.3 1134 <NA>
## 6 William K.L. Dickson <NA> NA 5.9 53 English
## Country Awards rtomRating rtomMeter rtomVotes Fresh Rotten rtomUserMeter
## 1 USA <NA> NA NA NA NA NA NA
## 2 France <NA> NA NA NA NA NA NA
## 3 France <NA> NA NA NA NA NA NA
## 4 France <NA> NA NA NA NA NA 100
## 5 USA 1 win. NA NA NA NA NA 32
## 6 USA <NA> NA NA NA NA NA NA
## rtomUserRating rtomUserVotes BoxOffice imdbRatingCatagory Genre_1
## 1 NA NA <NA> Average Documentary
## 2 NA NA <NA> Average Animation
## 3 NA NA <NA> Average Animation
## 4 2.8 216 <NA> Average Animation
## 5 3.0 184 <NA> Average Short
## 6 NA NA <NA> Average Short
## Genre_2 Genre_3
## 1 Short <NA>
## 2 Short <NA>
## 3 Comedy Short
## 4 Short <NA>
## 5 <NA> <NA>
## 6 <NA> <NA>We could also refer to the entire string by name but that would be tedious. Instead let’s use the subset function. We can combine that with the through statement we learned earlier (:) to grab a range. NOTE: This doesn’t count up the numbers after the _ but selects the Genre_4 and Genre_8 and everything that is between the two of them in the dataset. Even things that don’t have Genre in the title. We will learn more later about how to select things that are counting up.
Subset works by giving a data location, then a logical statement for selecting rows, then select which tells which columns to use. If we give it no logical statement it assumes all rows and if give no column statements it assums all columns.
movies <- subset(movies, select = -(Genre_4:Genre_8))Typical Genres
table(movies$Genre_1)##
## Action Adult Adventure Animation Biography Comedy
## 25638 16260 10160 25101 4134 74449
## Crime Documentary Drama Family Fantasy Film-Noir
## 13490 86989 81578 7988 1997 36
## Game-Show History Horror Music Musical Mystery
## 2101 1120 9673 7543 3126 2476
## News Reality-TV Romance Sci-Fi Short Sport
## 1576 5004 3347 2118 110902 2553
## Talk-Show Thriller War Western
## 2726 4885 763 3352Subset is a very useful command that we will be using a bit in this class. We can easily use it to replicate a few of the logical statements we made earlier with a bit more ease.
Like selecting very long movies Before we used: movies[which(movies$Runtime > 600), c(“Title”, “imdbRating”)]
head(subset(movies, Runtime > 600, select = c("Title", "imdbRating")))## Title imdbRating
## 51836 Semnadtsat mgnoveniy vesny 9.1
## 51860 War & Peace 8.2
## 52050 Chlopi 7.2
## 55075 Noce i dnie 7.4
## 55513 I, Claudius 9.1
## 55784 How Yukong Moved the Mountains 7.2We can embed that subset into a statement to select rows for things like graphing. In this case we subset the data then use the $ to create a vector of just imdbRatings.
hist(subset(movies, Runtime > 600)$imdbRating,
main = "Histogram of IMDB Ratings for movies Over 600 Minutes",
xlab = "IMDB ratings", xlim=c(1, 10), breaks = 10)Now that we are experts on splitting and subsetting data let’s go ahead and tackle a couple of other variables. Why don’t you try and work through Language and Country yourselves before following along.
head(table(movies$Language))##
## Abkhazian
## 2
## Aboriginal
## 16
## Aboriginal, English
## 5
## Aboriginal, English, French, Portuguese
## 1
## Aboriginal, Japanese, Hokkien
## 1
## Aboriginal, Polynesian, English
## 1head(table(movies$Country))##
## Afghanistan Afghanistan, France
## 21 2
## Afghanistan, France, Germany, UK Afghanistan, Iran
## 1 1
## Afghanistan, Ireland Afghanistan, Ireland, Japan
## 1 1Like before these are long strings seperated by commas. Since we still have splitstackshape loaded I will use that again. One of the positives is that you can pass a character vector to cSplit to split multiple variables simultaneously.
movies <- cSplit(movies, c("Language","Country"), sep=",")
setDF(movies)
summary(movies)## Title Year Appropriate Runtime
## Length:548010 Length:548010 Length:548010 Min. : 0.15
## Class :character Class :character Class :character 1st Qu.: 19.00
## Mode :character Mode :character Mode :character Median : 60.00
## Mean : 61.10
## 3rd Qu.: 91.00
## Max. :964.00
## NA's :76827
## Released Director Writer
## Min. :1888-10-14 Length:548010 Length:548010
## 1st Qu.:1982-12-03 Class :character Class :character
## Median :2004-09-23 Mode :character Mode :character
## Mean :1993-04-16
## 3rd Qu.:2010-06-13
## Max. :2022-01-01
## NA's :66665
## Metacritic imdbRating imdbVotes Awards
## Min. : 1.0 Min. : 1.00 Min. : 5 Length:548010
## 1st Qu.: 45.0 1st Qu.: 5.60 1st Qu.: 11 Class :character
## Median : 58.0 Median : 6.60 Median : 29 Mode :character
## Mean : 56.5 Mean : 6.38 Mean : 1614
## 3rd Qu.: 69.0 3rd Qu.: 7.30 3rd Qu.: 127
## Max. :100.0 Max. :10.00 Max. :1465210
## NA's :537880 NA's :197639 NA's :197640
## rtomRating rtomMeter rtomVotes Fresh
## Min. : 0.0 Min. : 0.0 Min. : 1.0 Min. : 0.0
## 1st Qu.: 5.0 1st Qu.: 40.0 1st Qu.: 6.0 1st Qu.: 3.0
## Median : 6.2 Median : 67.0 Median : 14.0 Median : 8.0
## Mean : 6.0 Mean : 61.6 Mean : 34.8 Mean : 21.6
## 3rd Qu.: 7.1 3rd Qu.: 86.0 3rd Qu.: 39.0 3rd Qu.: 24.0
## Max. :10.0 Max. :100.0 Max. :328.0 Max. :311.0
## NA's :529206 NA's :529017 NA's :526140 NA's :526140
## Rotten rtomUserMeter rtomUserRating rtomUserVotes
## Min. : 0.0 Min. : 0.0 Min. :0.0 Min. : 0
## 1st Qu.: 1.0 1st Qu.: 34.0 1st Qu.:2.2 1st Qu.: 6
## Median : 4.0 Median : 58.0 Median :3.1 Median : 74
## Mean : 13.2 Mean : 55.8 Mean :2.6 Mean : 25271
## 3rd Qu.: 12.0 3rd Qu.: 79.0 3rd Qu.:3.6 3rd Qu.: 512
## Max. :195.0 Max. :100.0 Max. :5.0 Max. :35791395
## NA's :526140 NA's :485854 NA's :455274 NA's :455131
## BoxOffice imdbRatingCatagory Genre_1
## Length:548010 Bad : 21960 Short :110902
## Class :character Average:289971 Documentary: 86989
## Mode :character Good : 38440 Drama : 81578
## NA's :197639 Comedy : 74449
## Action : 25638
## (Other) :131529
## NA's : 36925
## Genre_2 Genre_3 Language_01 Language_02
## Drama : 62760 Drama : 15207 English :274133 English: 7695
## Short : 46557 Romance : 11662 German : 28684 French : 3217
## Comedy : 36044 Thriller: 9668 Spanish : 27610 Spanish: 2834
## Romance: 14718 Comedy : 7354 French : 26076 German : 2366
## Crime : 9770 Family : 6791 Japanese: 18533 Tagalog: 1759
## (Other):102559 (Other) : 50743 (Other) :102686 (Other): 14433
## NA's :275602 NA's :446585 NA's : 70288 NA's :515706
## Language_03 Language_04 Language_05 Language_06
## English: 1881 English: 381 English: 112 German : 33
## French : 969 French : 307 French : 99 English: 32
## German : 828 German : 270 Spanish: 96 Spanish: 26
## Spanish: 590 Spanish: 203 German : 85 Italian: 24
## Italian: 443 Italian: 191 Italian: 75 Russian: 24
## (Other): 4148 (Other): 1525 (Other): 620 (Other): 218
## NA's :539151 NA's :545133 NA's :546923 NA's :547653
## Language_07 Language_08 Language_09
## Greek : 17 Hebrew : 10 Icelandic : 9
## Russian : 11 Portuguese: 7 Spanish : 5
## Portuguese: 10 Italian : 6 Italian : 4
## Polish : 9 Norwegian : 6 Russian : 4
## English : 7 Spanish : 6 Serbo-Croatian: 4
## (Other) : 110 (Other) : 66 (Other) : 43
## NA's :547846 NA's :547909 NA's :547941
## Language_10 Language_11 Language_12
## Italian : 9 Norwegian : 8 Portuguese: 5
## Portuguese: 6 Spanish : 5 Spanish : 4
## Spanish : 6 Swedish : 5 French : 3
## Polish : 4 Portuguese : 3 Polish : 3
## Danish : 3 Serbo-Croatian: 3 Italian : 2
## (Other) : 25 (Other) : 21 (Other) : 17
## NA's :547957 NA's :547965 NA's :547976
## Language_13 Language_14 Language_15
## Portuguese : 6 Serbo-Croatian: 4 Spanish : 5
## Serbo-Croatian: 4 Spanish : 4 Serbo-Croatian: 3
## Swedish : 3 Turkish : 4 Swedish : 3
## Raeto-Romance : 2 Slovenian : 3 German : 1
## Slovak : 2 Romanian : 2 Hebrew : 1
## (Other) : 9 (Other) : 7 (Other) : 4
## NA's :547984 NA's :547986 NA's :547993
## Language_16 Language_17 Language_18 Language_19
## Swedish : 5 Turkish: 5 Bengali : 1 Maltese: 1
## Turkish : 3 Spanish: 2 Quechua : 1 Russian: 1
## Slovenian: 2 Arabic : 1 Spanish : 2 Swedish: 1
## Spanish : 2 Finnish: 1 Swedish : 2 Turkish: 2
## Arabic : 1 Klingon: 1 Turkish : 1 Uzbek : 1
## (Other) : 3 (Other): 3 Ukrainian: 1 NA's :548004
## NA's :547994 NA's :547997 NA's :548002
## Language_20 Language_21 Language_22 Language_23
## Dutch : 1 Mode:logical Mode:logical Mode:logical
## Turkish: 1 NA's:548010 NA's:548010 NA's:548010
## NA's :548008
##
##
##
##
## Language_24 Language_25 Language_26 Language_27
## Mode:logical Mode:logical Mode:logical Mode:logical
## NA's:548010 NA's:548010 NA's:548010 NA's:548010
##
##
##
##
##
## Country_01 Country_02 Country_03 Country_04
## USA :202753 USA : 6556 France : 949 France : 236
## UK : 42264 France : 4579 USA : 851 USA : 213
## France : 25468 UK : 2689 Germany: 732 Germany: 190
## Canada : 20137 Canada : 2526 UK : 653 UK : 179
## Japan : 20082 Germany: 2389 Italy : 449 Italy : 144
## (Other):189691 (Other): 20353 (Other): 5030 (Other): 1694
## NA's : 47615 NA's :508918 NA's :539346 NA's :545354
## Country_05 Country_06 Country_07
## France : 83 Germany: 34 France : 13
## USA : 83 USA : 34 Germany : 13
## UK : 77 France : 29 Netherlands: 12
## Germany: 64 Italy : 26 UK : 11
## Spain : 36 UK : 23 USA : 11
## (Other): 664 (Other): 265 (Other) : 171
## NA's :547003 NA's :547599 NA's :547779
## Country_08 Country_09 Country_10
## Germany : 9 France : 8 UK : 4
## Austria : 6 Canada : 5 Australia: 3
## France : 6 Italy : 5 Belgium : 3
## Switzerland: 6 Belgium: 4 China : 3
## UK : 6 UK : 4 Israel : 3
## (Other) : 123 (Other): 79 (Other) : 55
## NA's :547854 NA's :547905 NA's :547939
## Country_11 Country_12 Country_13 Country_14
## Hungary : 4 Germany: 3 France : 5 Ecuador: 2
## Italy : 4 China : 2 India : 2 Estonia: 2
## South Africa: 4 Egypt : 2 Israel : 2 Germany: 2
## Finland : 3 Ireland: 2 Mexico : 2 Russia : 2
## Canada : 2 Italy : 2 UK : 2 Brazil : 1
## (Other) : 40 (Other): 27 (Other): 16 (Other): 14
## NA's :547953 NA's :547972 NA's :547981 NA's :547987
## Country_15 Country_16 Country_17
## Denmark: 3 Austria : 2 Croatia : 2
## Israel : 3 Czech Republic: 2 India : 2
## Georgia: 2 Congo : 1 China : 1
## Japan : 2 Côte d'Ivoire : 1 Czech Republic : 1
## China : 1 Ethiopia : 1 Dominican Republic: 1
## (Other): 9 (Other) : 11 (Other) : 7
## NA's :547990 NA's :547992 NA's :547996
## Country_18 Country_19 Country_20
## Bulgaria: 2 Belgium : 3 Austria : 2
## Cambodia: 2 Bangladesh : 1 Argentina: 1
## Brazil : 1 Bosnia and Herzegovina: 1 Australia: 1
## Croatia : 1 Brazil : 1 Belgium : 1
## Germany : 1 France : 1 Bolivia : 1
## (Other) : 6 (Other) : 6 (Other) : 7
## NA's :547997 NA's :547997 NA's :547997
## Country_21 Country_22 Country_23
## Albania : 1 Australia: 1 Argentina : 1
## Bhutan : 1 Canada : 1 Belgium : 1
## Chile : 1 France : 1 Egypt : 1
## India : 1 Russia : 1 South Africa: 1
## Philippines: 1 Slovenia : 1 Spain : 1
## (Other) : 4 (Other) : 2 (Other) : 2
## NA's :548001 NA's :548003 NA's :548003
## Country_24 Country_25 Country_26
## Australia : 1 Antarctica: 1 Cambodia: 1
## Czech Republic: 1 China : 1 NA's :548009
## Sweden : 1 Turkey : 1
## Syria : 1 UK : 1
## Uruguay : 1 NA's :548006
## NA's :548005
##
## Country_27
## Australia: 1
## NA's :548009
##
##
##
##
## Like before we will also need to deal with all these extra columns. Many of which don’t have much information contained within.
movies <- subset(movies, select = -(Language_04:Language_27))
movies <- subset(movies, select = -(Country_04:Country_27))Now that we have divided up language and country why don’t we see how many Turkish movies Are in this list. We also want to find out something about how good the movies are so let’s select movies that don’t have missing data for IMDb Ratings. Here we can combine what we have learned before to create a powerful search of the data to create a subset.
TurkishMovies <- subset(movies, Country_01=="Turkey" &
Language_01=="Turkish" &
!is.na(imdbRating),
select=c("Title", "imdbRating"))
head(TurkishMovies, n = 10)## Title imdbRating
## 13528 Aysel: Batakli damin kizi 7.1
## 45280 O Beautiful Istanbul 8.2
## 45503 Dry Summer 8.0
## 45856 Atesli delikanli 5.2
## 49119 Hope 8.2
## 49374 Little Ayse and the Magic Dwarfs in the Land of Dreams 5.0
## 50333 Umutsuzlar 7.3
## 50583 Agit 7.1
## 51942 Baba 7.3
## 52237 Gelin 7.8So what are the best and the worst Turkish movies? We can use order to arrange our dataset and head to display just the top and tail to display the bottom
TurkishMovies <- TurkishMovies[order(-TurkishMovies$imdbRating, TurkishMovies$Title), ]
head(TurkishMovies)## Title imdbRating
## 153141 The Chaos Class 9.5
## 480119 CM101MMXI Fundamentals 9.4
## 480973 CM101MMXI Fundamentals 9.4
## 377955 C.M.Y.L.M.Z. 9.3
## 533766 Kardes Payi 9.3
## 213335 Bir Tat Bir Doku 9.2tail(TurkishMovies)## Title imdbRating
## 461030 Hep ezildim 1.8
## 410372 Kahkaha Marketi 1.8
## 254248 Keloglan vs. the Black Prince 1.8
## 329286 Super Agent K9 1.4
## 298894 Yildizlar savasi 1.2
## 529115 Tersine 1.0It seems our dataset contains at least a few duplicated rows. For instance here we can see CM101MMXI Fundamentals seems to be so good it was included twice. We can verify that is in fact a duplicate by looking at it.
movies[c(480119, 480973), ]## Title Year Appropriate Runtime Released
## 480119 CM101MMXI Fundamentals 2013 <NA> 139 2013-01-03
## 480973 CM101MMXI Fundamentals 2013 <NA> 139 2013-01-03
## Director Writer Metacritic imdbRating imdbVotes Awards
## 480119 Murat Dundar Cem Yilmaz NA 9.4 32992 <NA>
## 480973 Murat Dundar Cem Yilmaz NA 9.4 32161 <NA>
## rtomRating rtomMeter rtomVotes Fresh Rotten rtomUserMeter
## 480119 NA NA NA NA NA NA
## 480973 NA NA NA NA NA NA
## rtomUserRating rtomUserVotes BoxOffice imdbRatingCatagory
## 480119 NA NA <NA> Good
## 480973 NA NA <NA> Good
## Genre_1 Genre_2 Genre_3 Language_01 Language_02 Language_03
## 480119 Documentary Comedy <NA> Turkish <NA> <NA>
## 480973 Documentary Comedy <NA> Turkish <NA> <NA>
## Country_01 Country_02 Country_03
## 480119 Turkey <NA> <NA>
## 480973 Turkey <NA> <NA>Finding duplicate rows can be tedious in a small dataset. In a dataset with over 500k observations it’s impossible to do manually. R has a couple of ways to identify unique and duplicate cases.
The easiest is duplicated(dataframe) which looks for two rows that are exactly the same. This will identify many duplicates but not ones with even superficial differences. For example, CM101MMXI Fundamentals has different numbers of votes in the imdbVotes variable so it wouldn’t be identified as a duplicate. It will also assume that the first row in the dataset is the original unless fromLast = TRUE. Finally, it is not the fastest operation. We can have it run a bit quicker by getting rid of movies that have the same Title but it will remove movies that are actually different that happen to have the same title. How you manage duplicates is up to you, for the purposes of this dataset a conservative approach is best since it’s likely many movies share a title.
We can save this as two data frames. One with the duplicated movies and the second is our movies dataframe with the duplicates removed.
duplicated_movies <- movies[duplicated(movies), ]
movies <- movies[!duplicated(movies), ]Exporting a File
Alright now let’s export this file so we can use it in later projects. It’s good data management to create new files after modifying your data so you can go back to previous versions or to store each iteration of your data in a new folder. Always preserve your original data without modifications. You have all the syntax here in R so you can easily re-run a syntax file partially to correct errors. It’s never that easy to recover missing, destroyed, or incorrectly modified data.
New Folder Method
At the beginning on the lesson we created a folder and set our working directory to that folder. Then we downloaded our file and called it movies_orig.RData. As long as we don’t overwrite that file we will have our original data. I prefer this method myself because I like folder based organization. Usually, I name my folders with the project name followed by the date I am working.
save(data, list of objects to save, file to save them in)
For example, we could save everything into an RData file with:
save(movies, file="movies.RData")or as a csv with
write.table(movies, file="movies.csv", sep=",")If you are going to continue using R I recommend keeping files in RData it’s faster and smaller.
file.info(c("movies.csv", "movies.RData"))## size isdir mode mtime ctime
## movies.csv 107340331 FALSE 666 2015-09-10 12:10:46 2015-09-09 15:12:51
## movies.RData 25541925 FALSE 666 2015-09-10 12:10:27 2015-09-09 15:12:43
## atime exe
## movies.csv 2015-09-09 15:12:51 no
## movies.RData 2015-09-09 15:12:43 noAutomatic Naming of Data Files
We can use R to automatically generate these saves in an easy to preserve format. We combine the paste which we have used before with the same command and a new command Sys.Date() which looks to your computer to find what today’s date is. This can be modified to include the hour, minute, or second to make completely sure every time you save a file it generates a new one. I find that having the day in front of the file is frequently good enough.
save(movies, file = paste(Sys.Date(), "movies.RData", sep=" "))
Let’s test out our knowledge of cleaning a dataset with lab 3. You can download it here: https://docs.google.com/document/d/17MFukYodljz6nIdgJIvS-G_g6B11nHcvZmsf4yhiP4M/edit?usp=sharing
Answers: https://drive.google.com/file/d/0BzzRhb-koTrLNWJsT2N4ZTAtU0U/view?usp=sharing
If you are having issues with the answers try downloading them and opening them in your browser of choice.